UMI/Barcode designs
Following schematic explains how Iso-Seq supports single-cell tags:
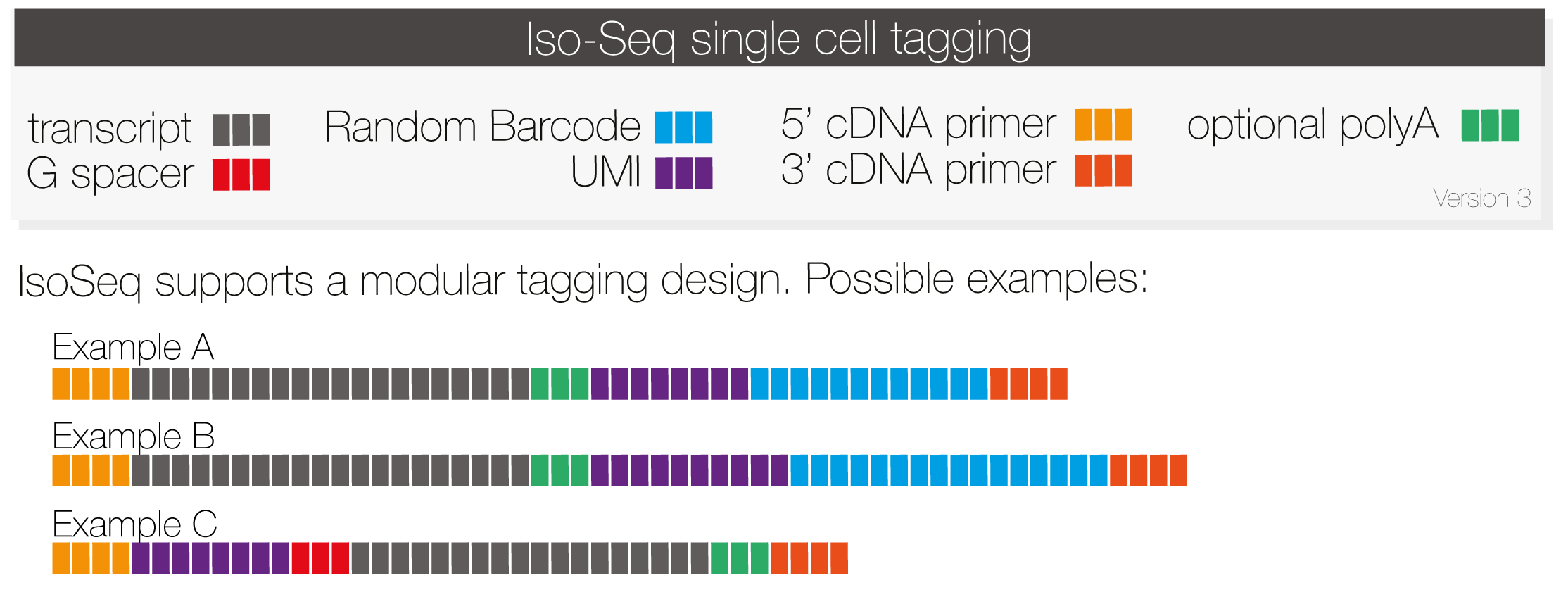
The tool of choice to clip tags (cell barcode, UMI, Gs), is isoseq tag. It supports design presets and custom experimental designs. Tags are abbreviated with a single character with optional length.
T indicates the transcript position and is mandatory. It is the anchor to determine if tags are located on the 3’ or 5’ side.
U as in UMI must be preceeded by the length of the UMI.
B as in cell barcode must be preceed by the length of the cell barcode.
G is a special PacBio 5’ UMI tag with a GGG suffix.
X must be must be preceeded by the length of an extra sequence to clip such as a TSO sequence.
Let’s explain by example, the Example A design of 8bp UMI 5’ and 12bp barcode 5’ can be specified with this string:
T-8U-12B
Tags are concatenated with hypens.
Similarly, the Example Bdesign:
T-10U-16B
The Example C design for a 8bp UMI 5’ with a fixed 3G UMI is:
8G-T
If you have a custom experiment, combine any of those tags uniquely, for example the a 6bp UMI 5’ and a 13bp barcode 3’:
6U-T-13U